
Abhay Kotecha
@AbhayKot
Structure Biologist, CryoEM expert, Atomic blobologist; DPhil @OxfordUni, scientist @ThermoFisher. also on Mastodon @[email protected]
ID:797106539421192193
11-11-2016 15:59:43
581 Tweets
1,9K Followers
553 Following


Great work by all involved at EMBL-EBI, EMDB - EMPIAR @EBI & BioImage Archive. Exciting dataset from
Abhay Kotecha team Microscopy & Spectroscopy. Important step to be able to visualise in this way. Foundational technology for the future😎
1800+ tomos of everyone’s favorite #Planimal 🌱 are online to browse in molecular detail. We hope this will be a resource for methods development and new biological discoveries. Shout out to Abhay Kotecha & team, Max Planck Institute of Biochemistry (MPIB), and our Biozentrum, University of Basel group (Ricardo🙏). Huge team effort!🙌

Excited to share our cryo-ET dataset: 1800+ tomos from 300+lamellae. Huge team work from team Ben Engel, team Max Planck Institute of Biochemistry (MPIB) & my team Microscopy & Spectroscopy + team EMDB - EMPIAR @EBI. We hope resources like this will be useful & push the frontier in cellular structure biology🚀 #CryoET

Victoria Cushing Simak Ali Abhay Kotecha Basil Greber and colleagues determine high-resolution cryo-EM structures of the CDK-activating kinase to establish a methodological framework for the use of cryo-EM in structure-based drug design.
nature.com/articles/s4146…



Further evidence of high-resolution capabilities at 100 keV with a side-entry cryo holder using the Gatan Microscopy Alpine detector. It even performed better than the K3 on a small sample - Great collab with colleagues in Verba lab UC San Francisco biorxiv.org/cgi/content/sh…
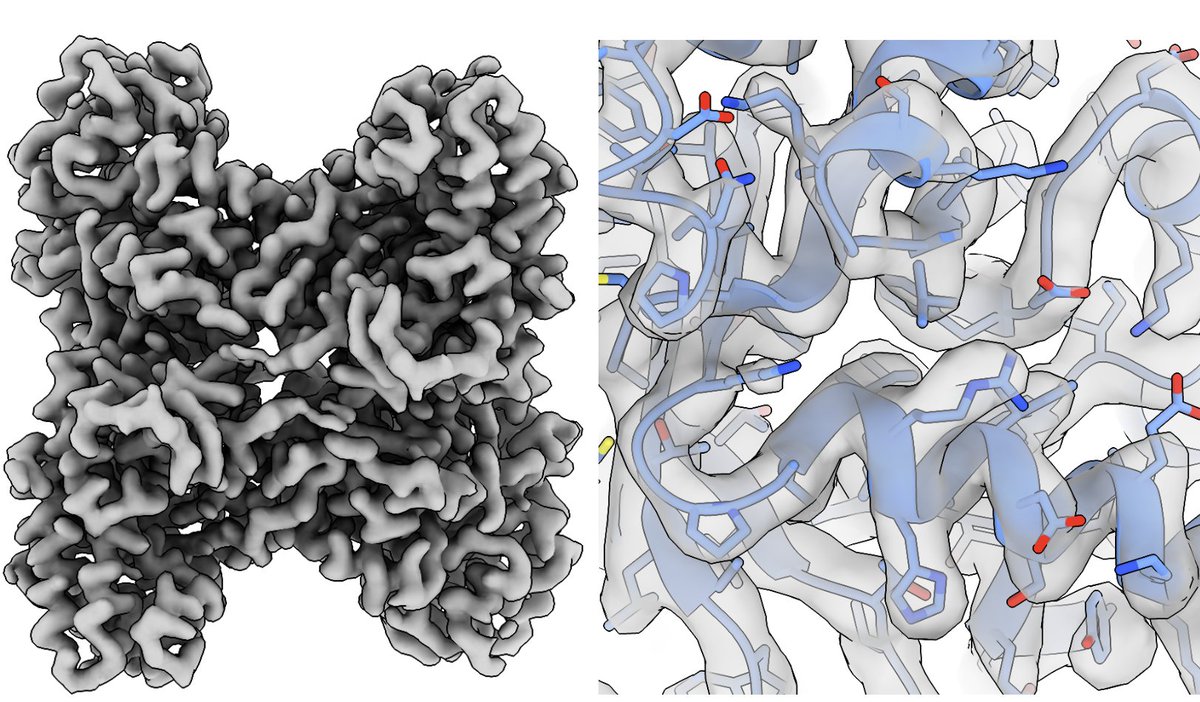

Thanks Basil Basil Greber! This is how it should always be. Wish that all would see & act this way. The obsession for the best resolution number needs to stop. Often misleading, overestimated and simply incorrect. Map quality, the region of interest and biological question matter!

Basil Greber Great! Because it is the map quality that matters, not the resolution number attached to it. 👍


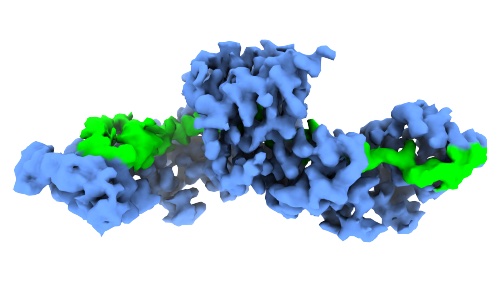
Our first 'lab paper' is out: In collaboration with Sandro Ataide/Yichen Zhong (University of Sydney) and Adrian Koh/Abhay Kotecha (TFS), Junjie determined the structure of the peptide binding domain of SRP68/72, the largest protein components of the eukaryotic SRP.


Would anyone in the hive perhaps have an idea what this could be? About 60 Angstroms across, probably made of 4x9 alpha-helices. Found in hexagonal arrays in mouse brain preps... (Would solve structure and use #ModelAngelo , but only have top views)


How small of a protein can you image with a Microscopy & Spectroscopy Tundra and Falcon C? Come join me and lingboyu tomorrow to find out! thermofisher.com/nl/en/home/glo…
Beautiful sample generously provided by Lander Lab



Welcome to the future of structural biology. Abhay Kotecha Thermo Fisher in collab with Ben Engel Florent Waltz unveil 5 mito complex structures in-situ from a single sample. And identify a potentially groundbreaking new membrane complex🔬🤩. Congrats everyone who contributed!



