New short paper from Joel Hellewell and Sam Horsfield and on making pangenome (specifically frequency estimates) from metagenome data:
biorxiv.org/cgi/content/sh…

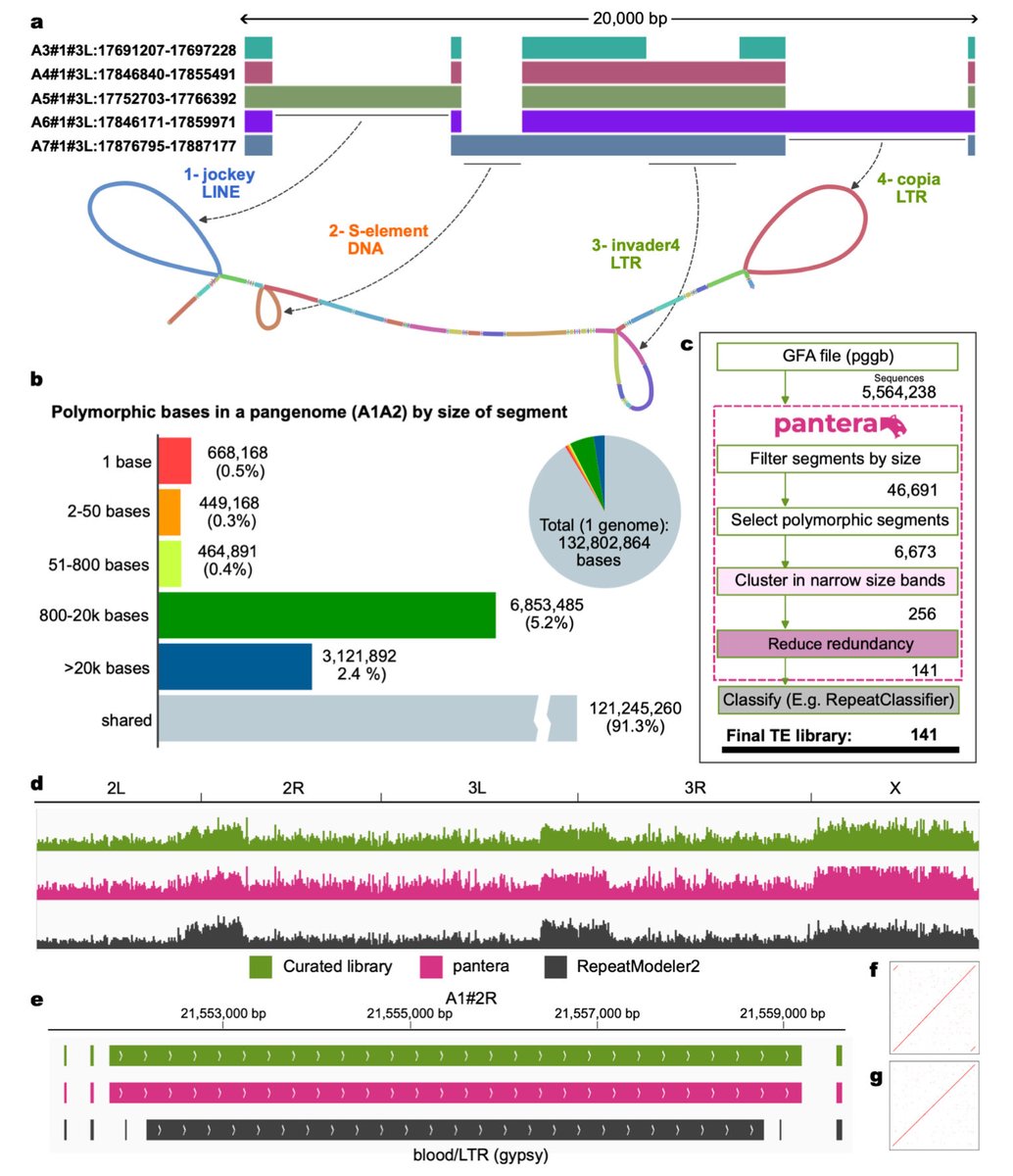
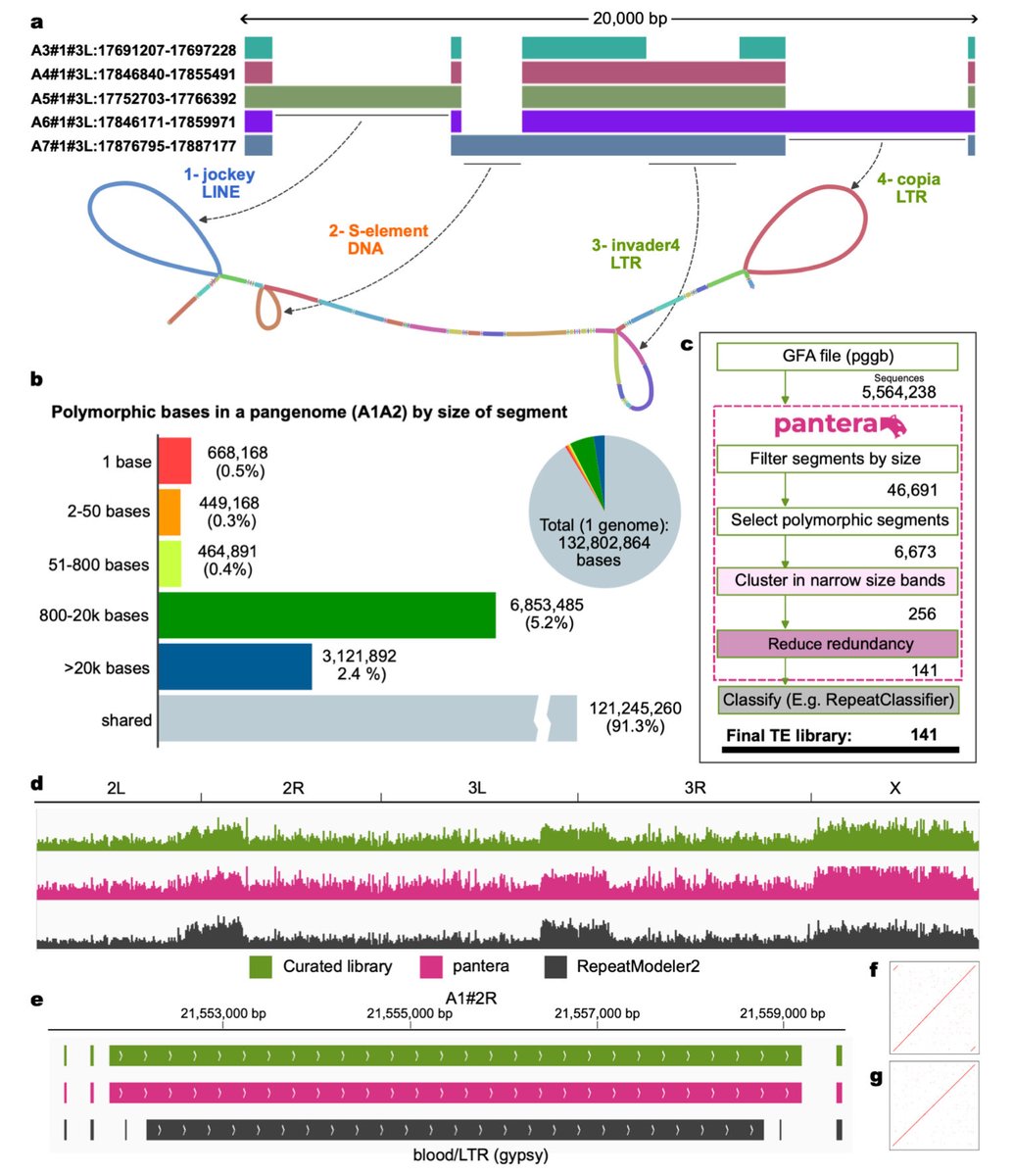
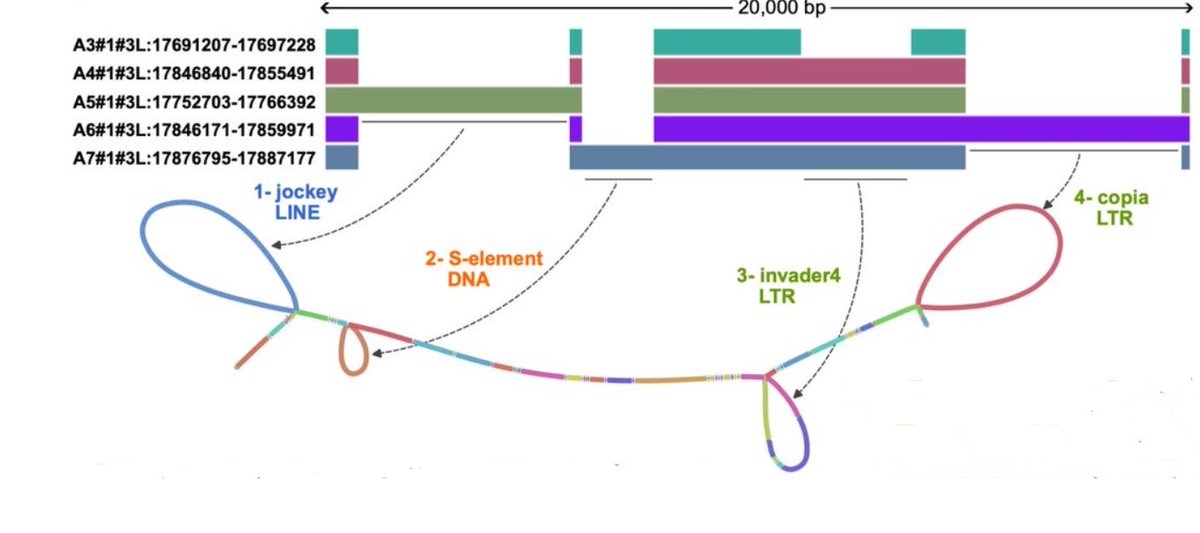
🧬🔍Very interesting to see how recently active #TransposableElements can be read out from pangenome graphs. Seems like a very good way forward to identify active TEs.
by pio Richard Durbin
doi.org/10.1101/2024.0…



Pangenome databases improve host removal and mycobacteria classification from clinical metagenomic data. #Pangenomes #HostRemoval #Metagenomics GigaScience
academic.oup.com/gigascience/ar…








Identification of transposable element families from pangenome polymorphisms biorxiv.org/cgi/content/sh… #biorxiv_bioinfo



Identification of transposable element families from pangenome polymorphisms. #TransposableElements #TEs #Pangenomes #Polymorphisms bioRxiv
biorxiv.org/content/10.110…