Axonal #TDP43 is a thing!
Proud to share our recent review written together with brilliant Topaz Altman and Perlson lab. We discuss the known and also the postulated functions of axonal and synaptic TDP43, and how these may relate to its peripheral toxicity in #ALS

Pleased to share our recent publication 'Multiple Copies of #microRNA Binding Sites in Long 3'UTR Variants Regulate Axonal Translation' achieved in a unique collaboration between Perlson lab and lab of Noam Shomron. More details in thread. #Axons #RNA
mdpi.com/2073-4409/12/2…

Here's an excellent review recently put out by the Perlson lab (Perlson lab, Ariel Ionescu, Topaz Altman) about axonal functions of TDP-43 and neuromuscular junction dysfunction in ALS. #endALS
…arneurodegeneration.biomedcentral.com/articles/10.11…

New publication for our ALS Review Series Collection!
'Looking for answers far away from the soma—the (un)known #axonal functions of TDP-43, and their contribution to early NMJ disruption in #ALS '
Ariel Ionescu Topaz Altman Perlson lab TAU Faculty of Medical & Health Sciences
bit.ly/3OH8Brs
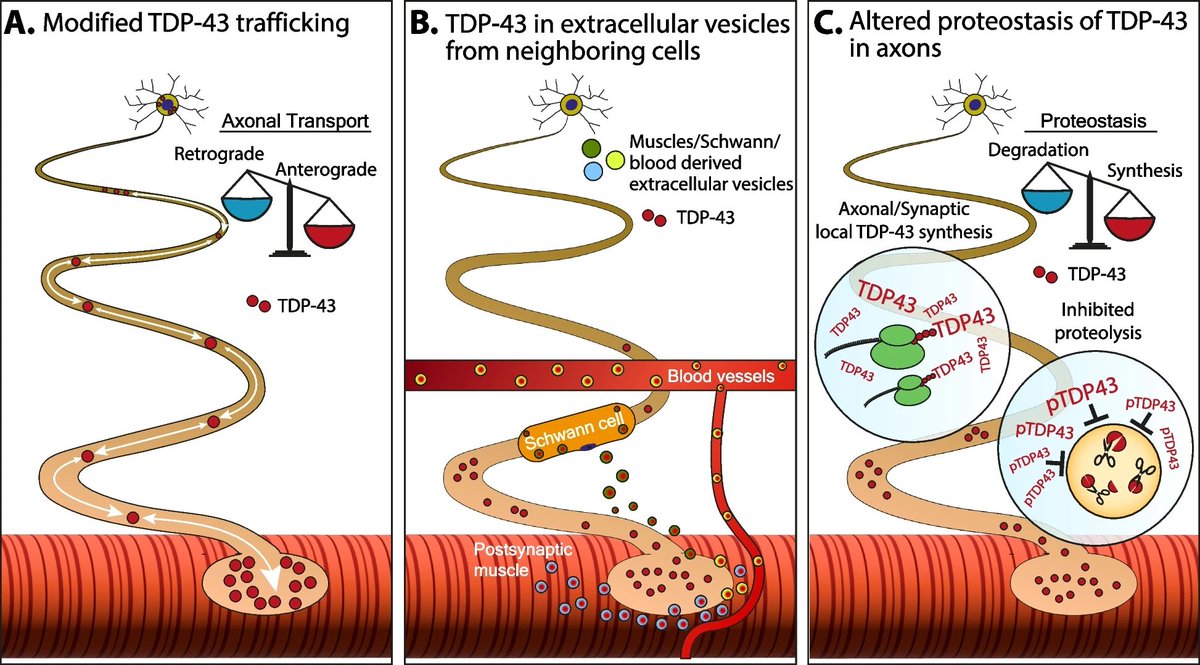












Ariel Ionescu Topaz Altman Perlson lab Congratulations to both of you Ariel Ionescu and Topaz Altman 🍾

Molecular Neurodegeneration Ariel Ionescu Topaz Altman Perlson lab TAU Faculty of Medical & Health Sciences Congratulations, Topaz, Ariel and Eran! On my reading list.